728x90
반응형
SMALL
Hodgkin Huxley
from brian2 import *
num_neurons = 100
duration = 2*second
# Parameters
area = 20000*umetre**2
Cm = 1*ufarad*cm**-2 * area
gl = 5e-5*siemens*cm**-2 * area
El = -65*mV
EK = -90*mV
ENa = 50*mV
g_na = 100*msiemens*cm**-2 * area
g_kd = 30*msiemens*cm**-2 * area
VT = -63*mV
# The model
eqs = Equations('''
dv/dt = (gl*(El-v) - g_na*(m*m*m)*h*(v-ENa) - g_kd*(n*n*n*n)*(v-EK) + I)/Cm : volt
dm/dt = 0.32*(mV**-1)*4*mV/exprel((13.*mV-v+VT)/(4*mV))/ms*(1-m)-0.28*(mV**-1)*5*mV/exprel((v-VT-40.*mV)/(5*mV))/ms*m : 1
dn/dt = 0.032*(mV**-1)*5*mV/exprel((15.*mV-v+VT)/(5*mV))/ms*(1.-n)-.5*exp((10.*mV-v+VT)/(40.*mV))/ms*n : 1
dh/dt = 0.128*exp((17.*mV-v+VT)/(18.*mV))/ms*(1.-h)-4./(1+exp((40.*mV-v+VT)/(5.*mV)))/ms*h : 1
I : amp
''')
# Threshold and refractoriness are only used for spike counting
group = NeuronGroup(num_neurons, eqs,
threshold='v > -40*mV',
refractory='v > -40*mV',
method='exponential_euler')
group.v = El
group.I = '0.7*nA * i / num_neurons'
monitor = SpikeMonitor(group)
run(duration)
plot(group.I/nA, monitor.count / duration)
xlabel('I (nA)')
ylabel('Firing rate (sp/s)')
show()

Adex (Adaptive exponential integrate and fire)
from brian2 import *
eqs = '''
dv/dt = -v/(10*ms) : volt
dvt/dt = (10*mV-vt)/(15*ms) : volt
'''
reset = '''
v = 0*mV
vt += 3*mV
'''
IF = NeuronGroup(1, model=eqs, reset=reset, threshold='v>vt',
method='exact')
IF.vt = 10*mV
PG = PoissonGroup(1, 500 * Hz)
C = Synapses(PG, IF, on_pre='v += 3*mV')
C.connect()
Mv = StateMonitor(IF, 'v', record=True)
Mvt = StateMonitor(IF, 'vt', record=True)
# Record the value of v when the threshold is crossed
M_crossings = SpikeMonitor(IF, variables='v')
run(2*second, report='text')
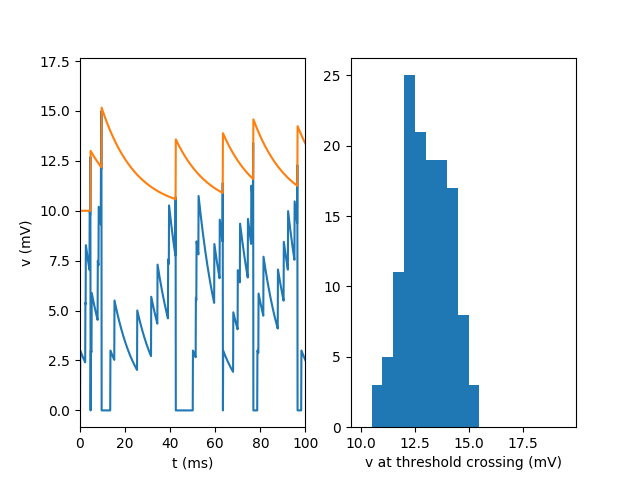
subplot(1, 2, 1)
plot(Mv.t / ms, Mv[0].v / mV)
plot(Mvt.t / ms, Mvt[0].vt / mV)
ylabel('v (mV)')
xlabel('t (ms)')
# zoom in on the first 100ms
xlim(0, 100)
subplot(1, 2, 2)
hist(M_crossings.v / mV, bins=np.arange(10, 20, 0.5))
xlabel('v at threshold crossing (mV)')
show()
728x90
반응형
LIST
'Natural Intelligence > Computational Neuroscience' 카테고리의 다른 글
| [Computational Neuroscience] 활동 전위 (Action potential) (0) | 2022.10.04 |
|---|---|
| [Computational Neuroscience] 신경의 흥분성 (Nerve Excitability) (0) | 2022.09.20 |
| [Brian] Introduction to Brian part 3 : Simulations (0) | 2022.02.14 |
| [Brian] Introduction to Brian part 2 : Synapses (0) | 2022.02.14 |
| [Brian] Introduction to Brian part 1 : Neurons (0) | 2022.02.11 |



