728x90
반응형
SMALL
COVID-19 Radiography

카타르 도하의 카타르 대학교와 방글라데시 다카 대학교의 연구진과 파키스탄, 말레이시아의 협력자들이 의사들과 협력하여 COVID-19 양성 사례에 대한 흉부 X선 영상 데이터베이스를 만들었다. 정상 및 바이러스성 폐렴 이미지, COVID-19, 정상 및 기타 폐 감염 데이터 세트로 구성되어 있다. 3616건의 COVID-19 양성 사례와 10,192건의 정상, 6012건의 Lung Opacity (비-COVID 폐 감염), 1345건의 바이러스성 폐렴 이미지 및 해당 폐 마스크로 데이터베이스를 확장했다.
캐글의 데이터셋을 다운로드하여 구글 드라이브에 업로드 후, 마운트를 진행한다.
import os
import glob
import numpy as np
import math
import numpy as np
import tensorflow as tf
from tensorflow.keras.layers import Dropout, Dense, GlobalAveragePooling2D
from tensorflow.keras.applications import MobileNet
from tensorflow.keras.models import Sequential
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from google.colab import drive
drive.mount('/content/gdrive/')import shutil
ROOT_DIR = '/content'
DATA_ROOT_DIR = os.path.join(ROOT_DIR, 'COVID-19_Radiography')
TRAIN_DATA_ROOT_DIR = os.path.join(DATA_ROOT_DIR, 'train')
TEST_DATA_ROOT_DIR = os.path.join(DATA_ROOT_DIR, 'test')
try:
dataset_path = '/content/gdrive/My Drive/Colab Notebooks/dataset'
shutil.copy(os.path.join(dataset_path, 'COVID-19_Radiography.zip'), '/content')
except Exception as err:
print(str(err))if os.path.exists(DATA_ROOT_DIR):
shutil.rmtree(DATA_ROOT_DIR)
print(DATA_ROOT_DIR + ' is removed.')# 압축파일 풀기
import zipfile
with zipfile.ZipFile(os.path.join(ROOT_DIR, 'COVID-19_Radiography.zip'), 'r') as target_file:
target_file.extractall(DATA_ROOT_DIR)
데이터 확인
# 전체 파일 확인 (디렉토리와 일부 파일 혼재하고 있음)
# isfile(), isdir() 사용하기 위해서는 full_path를 사용해야 하므로 os.listdir() 대신에 glob.glob() 사용
total_file_list = glob.glob(os.path.join(DATA_ROOT_DIR, 'COVID-19_Radiography_Dataset/*'))
print(total_file_list)
# 정답 리스트 추출 (디렉토리 이름이 정답)
label_name_list = [ file_name.split('/')[-1].strip() for file_name in total_file_list if os.path.isdir(file_name) == True ]
print(label_name_list)
# copytree 이용해서 정답이름/images 디렉토리를 train/정답 이름으로 복사
for label_name in label_name_list:
src_dir_path = os.path.join(DATA_ROOT_DIR, 'COVID-19_Radiography_Dataset'+'/'+label_name+'/images')
dst_dir_path = os.path.join(DATA_ROOT_DIR, 'train'+'/'+label_name)
try:
shutil.copytree(src_dir_path, dst_dir_path)
print(label_name+' copytree is done.')
except Exception as err:
print(str(err))['/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/COVID.metadata.xlsx', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Lung_Opacity.metadata.xlsx', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Normal', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/README.md.txt', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/COVID', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Normal.metadata.xlsx', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Viral Pneumonia.metadata.xlsx', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Viral Pneumonia', '/content/COVID-19_Radiography/COVID-19_Radiography_Dataset/Lung_Opacity']
['Normal', 'COVID', 'Viral Pneumonia', 'Lung_Opacity']
Normal copytree is done.
COVID copytree is done.
Viral Pneumonia copytree is done.
Lung_Opacity copytree is done.# train 정답 및 전체 데이터 개수 확인
train_label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
print(train_label_name_list)
for label_name in train_label_name_list:
print('train label : ', label_name,' => ', len(os.listdir(os.path.join(TRAIN_DATA_ROOT_DIR, label_name))))
print('='*54)['Normal', 'COVID', 'Viral Pneumonia', 'Lung_Opacity']
train label : Normal => 10192
train label : COVID => 3616
train label : Viral Pneumonia => 1345
train label : Lung_Opacity => 6012
======================================================# test dir 생성
if not os.path.exists(TEST_DATA_ROOT_DIR):
os.mkdir(TEST_DATA_ROOT_DIR)
print(TEST_DATA_ROOT_DIR + ' is created.')
else:
print(TEST_DATA_ROOT_DIR + ' already exists.')
# test dir 하위디렉토리에 정답 디렉토리 생성
for label_name in label_name_list:
if not os.path.exists(os.path.join(TEST_DATA_ROOT_DIR, label_name)):
os.mkdir(os.path.join(TEST_DATA_ROOT_DIR, label_name))
print(os.path.join(TEST_DATA_ROOT_DIR, label_name) + ' is created.')
else:
print(os.path.join(TEST_DATA_ROOT_DIR, label_name) + ' already exists.')/content/COVID-19_Radiography/test is created.
/content/COVID-19_Radiography/test/Normal is created.
/content/COVID-19_Radiography/test/COVID is created.
/content/COVID-19_Radiography/test/Viral Pneumonia is created.
/content/COVID-19_Radiography/test/Lung_Opacity is created.import random
# 파일 move 비율
MOVE_RATIO = 0.2 # train : test = 80 : 20, train 데이터 20% 데이터를 test 데이터로 사용
# 파일 move train_data_dir => test_data_dir
label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
for label_name in label_name_list:
# 파일 move 하기 위한 src_dir_path, dst_dir_path 설정
src_dir_path = os.path.join(TRAIN_DATA_ROOT_DIR,label_name)
dst_dir_path = os.path.join(TEST_DATA_ROOT_DIR,label_name)
train_data_file_list = os.listdir(src_dir_path)
print('='*47)
print('total [%s] data file nums => [%s]' % (label_name ,len(train_data_file_list)))
# data shuffle
random.shuffle(train_data_file_list)
print('train data shuffle is done.')
split_num = int(MOVE_RATIO*len(train_data_file_list))
print('split nums => ', split_num)
# extract test data from train data
test_data_file_list = train_data_file_list[0:split_num]
move_nums = 0
for test_data_file in test_data_file_list:
try:
shutil.move(os.path.join(src_dir_path, test_data_file),
os.path.join(dst_dir_path, test_data_file))
except Exception as err:
print(str(err))
move_nums = move_nums + 1
print('total move nums => ', move_nums)
print('='*47)===============================================
total [Normal] data file nums => [10192]
train data shuffle is done.
split nums => 2038
total move nums => 2038
===============================================
===============================================
total [COVID] data file nums => [3616]
train data shuffle is done.
split nums => 723
total move nums => 723
===============================================
===============================================
total [Viral Pneumonia] data file nums => [1345]
train data shuffle is done.
split nums => 269
total move nums => 269
===============================================
===============================================
total [Lung_Opacity] data file nums => [6012]
train data shuffle is done.
split nums => 1202
total move nums => 1202
===============================================# train 파일 개수 확인
label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
print(label_name_list)
for label_name in label_name_list:
label_dir = os.path.join(TRAIN_DATA_ROOT_DIR, label_name)
print('train label : ' + label_name + ' => ', len(os.listdir(os.path.join(TRAIN_DATA_ROOT_DIR, label_name))))
print('='*54)
# test 파일 개수 확인
label_name_list = os.listdir(TEST_DATA_ROOT_DIR)
print(label_name_list)
for label_name in label_name_list:
label_dir = os.path.join(TEST_DATA_ROOT_DIR, label_name)
print('test label : ' + label_name + ' => ', len(os.listdir(os.path.join(TEST_DATA_ROOT_DIR, label_name))))
print('='*54)['Normal', 'COVID', 'Viral Pneumonia', 'Lung_Opacity']
train label : Normal => 8154
train label : COVID => 2893
train label : Viral Pneumonia => 1076
train label : Lung_Opacity => 4810
======================================================
['Normal', 'COVID', 'Viral Pneumonia', 'Lung_Opacity']
test label : Normal => 2038
test label : COVID => 723
test label : Viral Pneumonia => 269
test label : Lung_Opacity => 1202
======================================================
데이터 전처리
IMG_WIDTH = 224
IMG_HEIGHT = 224
train_datagen = ImageDataGenerator(rescale=1./255, validation_split=0.2)
validation_data_gen = ImageDataGenerator(rescale=1./255, validation_split=0.2)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(TRAIN_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT),
subset='training')
validation_generator = validation_data_gen.flow_from_directory(TRAIN_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT),
subset='validation')
test_generator = test_datagen.flow_from_directory(TEST_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT))Found 13548 images belonging to 4 classes.
Found 3385 images belonging to 4 classes.
Found 4232 images belonging to 4 classes.print(train_generator.class_indices)
print(train_generator.num_classes){'COVID': 0, 'Lung_Opacity': 1, 'Normal': 2, 'Viral Pneumonia': 3}
4import matplotlib.pyplot as plt
IMG_NUMS = 16
image_data, image_label = train_generator.next()
data = image_data[:IMG_NUMS]
label = image_label[:IMG_NUMS]
class_dict = {0:'COVID', 1:'Lung_Opacity', 2:'Normal', 3:'Viral Pneumonia'}
plt.figure(figsize=(9,9))
for i in range(len(label)):
plt.subplot(4, 4, i+1)
plt.title(str(class_dict[label[i]]))
plt.xticks([]); plt.yticks([])
plt.imshow(data[i])
plt.tight_layout()
plt.show()
모델 생성
pre_trained_model = MobileNet(weights='imagenet', include_top=False, input_shape=(IMG_WIDTH,IMG_HEIGHT,3))
class_nums = train_generator.num_classes
model = Sequential()
model.add(pre_trained_model)
model.add(GlobalAveragePooling2D())
model.add(Dense(64,activation='relu'))
model.add(Dropout(0.5)) # 0.25 오버피팅 발생함
model.add(Dense(class_nums, activation='softmax'))
model.summary()Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
mobilenet_1.00_224 (Functio (None, 7, 7, 1024) 3228864
nal)
global_average_pooling2d (G (None, 1024) 0
lobalAveragePooling2D)
dense (Dense) (None, 64) 65600
dropout (Dropout) (None, 64) 0
dense_1 (Dense) (None, 4) 260
=================================================================
Total params: 3,294,724
Trainable params: 3,272,836
Non-trainable params: 21,888
_________________________________________________________________from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
from datetime import datetime
model.compile(loss='sparse_categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(2e-5), metrics=['acc'])
save_file_name = './COVID-19_Radiography_MobileNet_Colab.h5'
checkpoint = ModelCheckpoint(save_file_name, # file명을 지정
monitor='val_loss', # val_loss 값이 개선되었을때 호출
verbose=1, # 로그를 출력
save_best_only=True, # 가장 best 값만 저장
mode='auto' # auto는 알아서 best를 찾음. min/max
)
earlystopping = EarlyStopping(monitor='val_loss', # 모니터 기준 설정 (val loss)
patience=5, # 3회 Epoch동안 개선되지 않는다면 종료
verbose=1
)
start_time = datetime.now()
hist = model.fit(train_generator, epochs=20, validation_data=validation_generator)
end_time = datetime.now()
print('Elapsed Time => ', end_time-start_time)plt.plot(hist.history['acc'], label='train')
plt.plot(hist.history['val_acc'], label='validation')
plt.title('Accuracy Trend')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(loc='best')
plt.grid()
plt.show()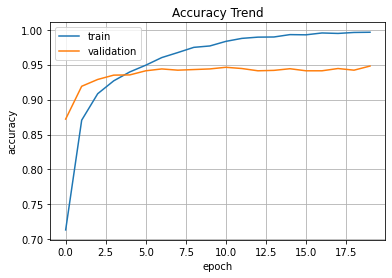
plt.plot(hist.history['loss'], label='train')
plt.plot(hist.history['val_loss'], label='validation')
plt.title('Loss Trend')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(loc='best')
plt.grid()
plt.show()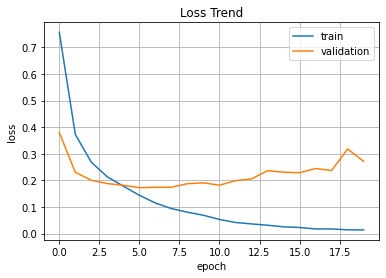
model.evaluate(test_generator)[0.34339311718940735, 0.9328922629356384]728x90
반응형
LIST
'Visual Intelligence > Image Deep Learning' 카테고리의 다른 글
| [시각 지능] TPU (Tensor Processing Unit) (0) | 2022.09.03 |
|---|---|
| [시각 지능] Coffee Classification (0) | 2022.08.28 |
| [시각 지능] Chest X-Ray Images (Pneumonia) (0) | 2022.08.27 |
| [시각 지능] Brain Tumor Classification (MRI) (1) | 2022.08.21 |
| [시각 지능] imgaug (0) | 2022.08.21 |



