728x90
반응형
SMALL
Brain Tumor Classification (MRI)

뇌종양은 어린이와 성인 사이에서 공격적인 질병 중 하나로 간주된다. 뇌종양은 모든 원발성 중추신경계 (CNS) 종양의 85~90%를 차지한다. 매년 약 11,700명이 뇌종양 진단을 받는다. 암성 뇌 또는 CNS 종양이 있는 사람들의 5년 생존율은 남성의 경우 약 34%, 여성의 경우 36%이다. 뇌종양은 양성종양, 악성종양, 뇌하수체종양 등으로 분류된다. 환자의 기대수명을 향상시키기 위해서는 적절한 치료와 계획, 정확한 진단이 이루어져야 한다. 뇌종양을 탐지하는 가장 좋은 방법은 자기공명영상촬영 (MRI)이다. 스캔을 통해 엄청난 양의 이미지 데이터가 생성된다. 이 이미지는 방사선 전문의가 검사한다.
캐글에서 Brain Tumor Classification (MRI) dataset을 add한다.
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.applications import MobileNet, Xception
from tensorflow.keras.layers import Flatten, Dense, Dropout, GlobalAveragePooling2D
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import os
ROOT_DIR = '../input/'
DATA_ROOT_DIR = os.path.join(ROOT_DIR, 'brain-tumor-classification-mri')
TRAIN_DATA_ROOT_DIR = os.path.join(DATA_ROOT_DIR, 'Training')
TEST_DATA_ROOT_DIR = os.path.join(DATA_ROOT_DIR, 'Testing')
# train 정답 및 전체 데이터 개수 확인
train_label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
print(train_label_name_list)
for label_name in train_label_name_list:
print('train label : ', label_name,' => ', len(os.listdir(os.path.join(TRAIN_DATA_ROOT_DIR, label_name))))
print('='*67)
# test 정답 및 전체 데이터 개수 확인
test_label_name_list = os.listdir(TEST_DATA_ROOT_DIR)
print(test_label_name_list)
for label_name in test_label_name_list:
print('test label : ', label_name, ' => ', len(os.listdir(os.path.join(TEST_DATA_ROOT_DIR, label_name))))
print('='*67)['no_tumor', 'pituitary_tumor', 'meningioma_tumor', 'glioma_tumor']
train label : no_tumor => 395
train label : pituitary_tumor => 827
train label : meningioma_tumor => 822
train label : glioma_tumor => 826
===================================================================
['no_tumor', 'pituitary_tumor', 'meningioma_tumor', 'glioma_tumor']
test label : no_tumor => 105
test label : pituitary_tumor => 74
test label : meningioma_tumor => 115
test label : glioma_tumor => 100
===================================================================
ImageDataGenerator
IMG_WIDTH = 224
IMG_HEIGHT = 224
train_datagen = ImageDataGenerator(rescale=1./255, validation_split=0.2)
validation_datagen = ImageDataGenerator(rescale=1./255, validation_split=0.2)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(TRAIN_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT),
subset='training')
validation_generator = train_datagen.flow_from_directory(TRAIN_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT),
subset='validation')
test_generator = test_datagen.flow_from_directory(TEST_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT))Found 2297 images belonging to 4 classes.
Found 573 images belonging to 4 classes.
Found 394 images belonging to 4 classes.print(train_generator.class_indices)
print(train_generator.num_classes){'glioma_tumor': 0, 'meningioma_tumor': 1, 'no_tumor': 2, 'pituitary_tumor': 3}
4IMG_NUMS = 16
image_data, label_data = train_generator.next()
data = image_data[:IMG_NUMS]
label = label_data[:IMG_NUMS]
print(data.shape, label.shape)
print(label)(16, 224, 224, 3) (16,)
[0. 0. 1. 1. 2. 1. 0. 3. 3. 0. 3. 2. 2. 3. 0. 3.]import matplotlib.pyplot as plt
class_dict = { 0:'glioma_tumor', 1:'meningioma_tumor', 2:'no_tumor', 3:'pituitary_tumor'}
plt.figure(figsize=(8, 8))
for i in range(len(label)):
plt.subplot(4, 4, i+1)
plt.title(str(class_dict[label[i]]))
plt.xticks([]); plt.yticks([])
plt.imshow(data[i])
plt.tight_layout()
plt.show()
Fine Tuning
pre_trained_model = MobileNet(weights='imagenet', include_top=False, input_shape=(IMG_WIDTH,IMG_HEIGHT,3))
class_nums = train_generator.num_classes # 정답 개수
model = Sequential()
model.add(pre_trained_model)
model.add(GlobalAveragePooling2D())
model.add(Dense(512,activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(128,activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(class_nums, activation='softmax'))
model.summary()Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
mobilenet_1.00_224 (Function (None, 7, 7, 1024) 3228864
_________________________________________________________________
global_average_pooling2d (Gl (None, 1024) 0
_________________________________________________________________
dense (Dense) (None, 512) 524800
_________________________________________________________________
dropout (Dropout) (None, 512) 0
_________________________________________________________________
dense_1 (Dense) (None, 128) 65664
_________________________________________________________________
dropout_1 (Dropout) (None, 128) 0
_________________________________________________________________
dense_2 (Dense) (None, 4) 516
=================================================================
Total params: 3,819,844
Trainable params: 3,797,956
Non-trainable params: 21,888
_________________________________________________________________from tensorflow.keras.callbacks import EarlyStopping
from datetime import datetime
model.compile(loss='sparse_categorical_crossentropy',
optimizer=tf.keras.optimizers.Adam(2e-5), metrics=['acc'])
earlystopping = EarlyStopping(monitor='val_loss', # 모니터 기준 설정 (val loss)
patience=5, # 5회 Epoch동안 개선되지 않는다면 종료
verbose=1)
start_time = datetime.now()
hist = model.fit(train_generator, epochs=30, validation_data=validation_generator)
end_time = datetime.now()
print('Elapsed Time => ', end_time-start_time)plt.plot(hist.history['acc'], label='train')
plt.plot(hist.history['val_acc'], label='validation')
plt.title('Accuracy Trend')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(loc='best')
plt.grid()
plt.show()
plt.plot(hist.history['loss'], label='train')
plt.plot(hist.history['val_loss'], label='validation')
plt.title('Loss Trend')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(loc='best')
plt.grid()
plt.show()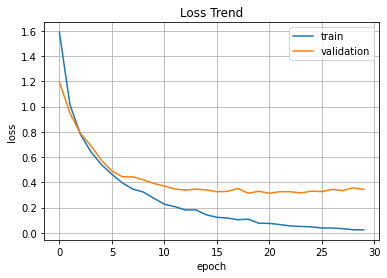
evaluate
model.evaluate(test_generator)[1.6366639137268066, 0.7360405921936035]
homogeneous를 위한 test data mix
test_label_name_list = os.listdir(TEST_DATA_ROOT_DIR)
print(test_label_name_list)
for label_name in test_label_name_list: # Testing 디렉토리 각각의 label에서 실행
test_path = os.path.join(TEST_DATA_ROOT_DIR, label_name)
test_file_list = os.listdir(test_path)
train_path = os.path.join(TRAIN_DATA_ROOT_DIR, label_name)
moved_num = 0
for test_image_file in test_file_list: # Testing 모든 data를 Training 디렉토리로 이동
# already exists error 방지
# 참고소스: https://stackoverflow.com/questions/31813504
shutil.move(os.path.join(test_path, test_image_file),
os.path.join(train_path, test_image_file))
moved_num = moved_num + 1
print(str(moved_num) + ' is moved into ' + label_name + ' (Testing => Training)')['no_tumor', 'pituitary_tumor', 'glioma_tumor', 'meningioma_tumor']
105 is moved into no_tumor (Testing => Training)
74 is moved into pituitary_tumor (Testing => Training)
100 is moved into glioma_tumor (Testing => Training)
115 is moved into meningioma_tumor (Testing => Training)# train 정답 및 전체 데이터 개수 확인
train_label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
print(train_label_name_list)
for label_name in train_label_name_list:
print('train label : ', label_name,' => ', len(os.listdir(os.path.join(TRAIN_DATA_ROOT_DIR, label_name))))
print('='*67)['no_tumor', 'pituitary_tumor', 'glioma_tumor', 'meningioma_tumor']
train label : no_tumor => 396
train label : pituitary_tumor => 901
train label : glioma_tumor => 926
train label : meningioma_tumor => 937# 파일 move 비율
MOVE_RATIO = 0.2 # train : test = 80 : 20, train 데이터 20% 데이터를 test 데이터로 사용# 파일 move train_data_dir => test_data_dir
label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
for label_name in label_name_list:
# 파일 move 하기 위한 src_dir_path, dst_dir_path 설정
src_dir_path = os.path.join(TRAIN_DATA_ROOT_DIR,label_name)
dst_dir_path = os.path.join(TEST_DATA_ROOT_DIR,label_name)
train_data_file_list = os.listdir(src_dir_path)
print('========================================================================')
print('total [%s] data file nums => [%s]' % (label_name ,len(train_data_file_list)))
# data shuffle
random.shuffle(train_data_file_list)
print('train data shuffle is done !!!')
split_num = int(MOVE_RATIO*len(train_data_file_list))
print('split nums => ', split_num)
# extract test data from train data
test_data_file_list = train_data_file_list[0:split_num]
move_nums = 0
for test_data_file in test_data_file_list:
try:
shutil.move(os.path.join(src_dir_path, test_data_file),
os.path.join(dst_dir_path, test_data_file))
except Exception as err:
print(str(err))
move_nums = move_nums + 1
print('total move nums => ', move_nums)
print('========================================================================')========================================================================
total [no_tumor] data file nums => [396]
train data shuffle is done !!!
split nums => 79
total move nums => 79
========================================================================
========================================================================
total [pituitary_tumor] data file nums => [901]
train data shuffle is done !!!
split nums => 180
total move nums => 180
========================================================================
========================================================================
total [glioma_tumor] data file nums => [926]
train data shuffle is done !!!
split nums => 185
total move nums => 185
========================================================================
========================================================================
total [meningioma_tumor] data file nums => [937]
train data shuffle is done !!!
split nums => 187
total move nums => 187
========================================================================# train 파일 개수 확인
label_name_list = os.listdir(TRAIN_DATA_ROOT_DIR)
print(label_name_list)
for label_name in label_name_list:
label_dir = os.path.join(TRAIN_DATA_ROOT_DIR, label_name)
print('train label : ' + label_name + ' => ', len(os.listdir(os.path.join(TRAIN_DATA_ROOT_DIR, label_name))))
print('=====================================================')
# test 파일 개수 확인
label_name_list = os.listdir(TEST_DATA_ROOT_DIR)
print(label_name_list)
for label_name in label_name_list:
label_dir = os.path.join(TEST_DATA_ROOT_DIR, label_name)
print('test label : ' + label_name + ' => ', len(os.listdir(os.path.join(TEST_DATA_ROOT_DIR, label_name))))
print('=====================================================')['no_tumor', 'pituitary_tumor', 'glioma_tumor', 'meningioma_tumor']
train label : no_tumor => 317
train label : pituitary_tumor => 721
train label : glioma_tumor => 741
train label : meningioma_tumor => 750
=====================================================
['no_tumor', 'pituitary_tumor', 'glioma_tumor', 'meningioma_tumor']
test label : no_tumor => 79
test label : pituitary_tumor => 180
test label : glioma_tumor => 185
test label : meningioma_tumor => 187
=====================================================IMG_WIDTH = 224
IMG_HEIGHT = 224
train_datagen = ImageDataGenerator(rescale=1./255)
test_datagen = ImageDataGenerator(rescale=1./255)
train_generator = train_datagen.flow_from_directory(TRAIN_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT))
test_generator = test_datagen.flow_from_directory(TEST_DATA_ROOT_DIR, batch_size=32,
color_mode='rgb', class_mode='sparse',
target_size=(IMG_WIDTH,IMG_HEIGHT))Found 2529 images belonging to 4 classes.
Found 631 images belonging to 4 classes.print(train_generator.class_indices)
print(train_generator.num_classes){'glioma_tumor': 0, 'meningioma_tumor': 1, 'no_tumor': 2, 'pituitary_tumor': 3}
4# 32개의 데이터와 label을 가져와서 16개 출력
IMG_NUMS = 16
image_data, label_data = train_generator.next()
data = image_data[:IMG_NUMS]
label = label_data[:IMG_NUMS]
print(data.shape, label.shape)
print(label)(32, 224, 224, 3) (32,)
[1. 3. 0. 0. 0. 3. 0. 1. 0. 0. 3. 2. 3. 1. 0. 3. 3. 1. 0. 1. 0. 0. 2. 1.
3. 1. 0. 0. 1. 3. 2. 2.]class_dict = { 0:'glioma_tumor', 1:'meningioma_tumor', 2:'no_tumor', 3:'pituitary_tumor'}
plt.figure(figsize=(8,8))
for i in range(len(label)):
plt.subplot(4, 4, i+1)
plt.title(str(class_dict[label[i]]))
plt.xticks([]); plt.yticks([])
plt.imshow(data[i])
plt.tight_layout()
plt.show()
hist = model.fit(train_generator, epochs=50, validation_data=test_generator, callbacks=[earlystopping])
model.evaluate(test_generator)[0.19745692610740662, 0.9461172819137573]
728x90
반응형
LIST
'Visual Intelligence > Image Deep Learning' 카테고리의 다른 글
| [시각 지능] COVID-19 Radiography (0) | 2022.08.27 |
|---|---|
| [시각 지능] Chest X-Ray Images (Pneumonia) (0) | 2022.08.27 |
| [시각 지능] imgaug (0) | 2022.08.21 |
| [시각 지능] Image Data Augmentation + Transfer Learning (0) | 2022.08.20 |
| [시각 지능] Image Data Augmentation (0) | 2022.08.20 |



