728x90
반응형
SMALL
import matplotlib
import pandas as pd
from matplotlib import pyplot as plt
df = pd.read_csv('./kaggle_diabetes.csv')
df.head()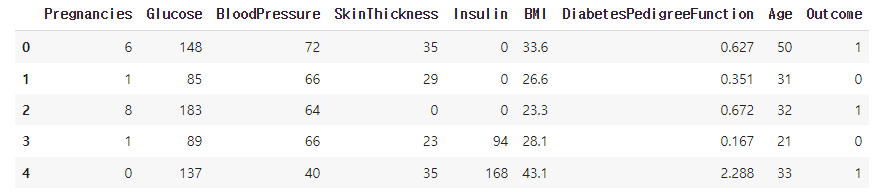
df.hist()
plt.tight_layout()
plt.show()
df['BloodPressure'].hist()
plt.tight_layout()
plt.show()
df.info()
df.isnull().sum()
for col in df.columns:
missing_rows = df.loc[df[col] == 0].shape[0]
print(col + ": " + str(missing_rows))
import numpy as np
# outlier 처리
df['Glucose'] = df['Glucose'].replace(0, np.nan)
df['BloodPressure'] = df['BloodPressure'].replace(0, np.nan)
df['SkinThickness'] = df['SkinThickness'].replace(0, np.nan)
df['Insulin'] = df['Insulin'].replace(0, np.nan)
df['BMI'] = df['BMI'].replace(0, np.nan)
# missing value 처리
df['Glucose'] = df['Glucose'].fillna(df['Glucose'].mean())
df['BloodPressure'] = df['BloodPressure'].fillna(df['BloodPressure'].mean())
df['SkinThickness'] = df['SkinThickness'].fillna(df['SkinThickness'].mean())
df['Insulin'] = df['Insulin'].fillna(df['Insulin'].mean())
df['BMI'] = df['BMI'].fillna(df['BMI'].mean())
for col in df.columns:
missing_rows = df.loc[df[col] == 0].shape[0]
print(col + ": " + str(missing_rows))
# raw 데이터프레임 보존
df_scaled = df
df_scaled.describe()# feature column, label column 추출 후 데이터프레임 생성
feature_df = df_scaled[df_scaled.columns.difference(['Outcome'])]
label_df = df_scaled['Outcome']
print(feature_df.shape, label_df.shape)
# pandas <=> numpy
feature_np = feature_df.to_numpy().astype('float32')
label_np = label_df.to_numpy().astype('float32')
print(feature_np.shape, label_np.shape)
s = np.arange(len(feature_np))
np.random.shuffle(s)
feature_np = feature_np[s]
label_np = label_np[s]
# train / test 데이터 분리
split = 0.15
test_num = int(split*len(label_np))
x_test = feature_np[0:test_num]
y_test = label_np[0:test_num]
x_train = feature_np[test_num:]
y_train = label_np[test_num:]
print(x_train.shape, y_train.shape)
print(x_test.shape, y_test.shape)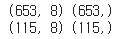
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Dropout
model = Sequential()
model.add(Dense(1, activation = 'sigmoid', input_shape = (8,)))
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=1e-3),
loss='binary_crossentropy', metrics=['accuracy'])
from datetime import datetime
start_time = datetime.now()
hist = model.fit(x_train, y_train, epochs=400, validation_data=(x_test, y_test), verbose=2)
end_time = datetime.now()
print('elapsed time => ', end_time - start_time)model.evaluate(x_test, y_test)4/4 [==============================] - 0s 10ms/step - loss: 2.8208 - accuracy: 0.7217
[2.820805549621582, 0.7217391133308411]import matplotlib.pyplot as plt
plt.title('loss trend')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.grid()
plt.plot(hist.history['loss'], label = 'train loss')
plt.plot(hist.history['val_loss'], label = 'validation loss')
plt.legend(loc = 'best')
plt.show()
plt.title('accuracy trend')
plt.xlabel('epochs')
plt.ylabel('loss')
plt.grid()
plt.plot(hist.history['accuracy'], label = 'train accuracy')
plt.plot(hist.history['val_accuracy'], label = 'validation accuracy')
plt.legend(loc = 'best')
plt.show()
표준화 (Standardization)

데이터의 피처 각각이 평 균이 0, 분산이 1인 가우시안 정규분포를 가진 값으 로 변환하는 작업을 표준화라고 한다. 실제 구현시에는 사이킷런의 StandardScaler를 사용해 표준화를 진행하는것이 일반적이다.
# 표준화 (Standardization)
from sklearn.preprocessing import StandardScaler
scale_cols = ['Pregnancies', 'Glucose', 'BloodPressure', 'SkinThickness',
'Insulin', 'BMI', 'DiabetesPedigreeFunction', 'Age']
scaler = StandardScaler()
df_std = scaler.fit_transform(df[scale_cols])
print(type(df_std))
df_scaled = pd.DataFrame(df_std, columns=scale_cols)
df_scaled['Outcome'] = df['Outcome'].values # raw 데이터프레임 보존
df_scaled.describe()feature_df = df_scaled[df_scaled.columns.difference(['Outcome'])]
label_df = df_scaled['Outcome']
print(feature_df.shape, label_df.shape)
# pandas <=> numpy
feature_np = feature_df.to_numpy().astype('float32')
label_np = label_df.to_numpy().astype('float32')
print(feature_np.shape, label_np.shape)
s = np.arange(len(feature_np))
np.random.shuffle(s)
feature_np = feature_np[s]
label_np = label_np[s]
# train / test 데이터 분리
split = 0.15
test_num = int(split*len(label_np))
x_test = feature_np[0:test_num]
y_test = label_np[0:test_num]
x_train = feature_np[test_num:]
y_train = label_np[test_num:]
print(x_train.shape, y_train.shape)
print(x_test.shape, y_test.shape)
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Dropout
model = Sequential()
model.add(Dense(1, activation = 'sigmoid', input_shape = (8,)))
model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=1e-3),
loss='binary_crossentropy', metrics=['accuracy'])
hist = model.fit(x_train, y_train, epochs=400, validation_data=(x_test, y_test), verbose=2)
model.evaluate(x_test, y_test)4/4 [==============================] - 0s 3ms/step - loss: 0.4877 - accuracy: 0.7217
[0.48767489194869995, 0.7217391133308411]

728x90
반응형
LIST
'AI-driven Methodology > Artificial Intelligence' 카테고리의 다른 글
| [AI] 와인 종류 예측 (0) | 2022.07.24 |
|---|---|
| [AI] 와인 품질 예측 (0) | 2022.07.24 |
| [AI] 분류 (Classification) (0) | 2022.07.23 |
| [AI] 다변수 선형 회귀 (0) | 2022.07.17 |
| [AI] Basic Architecture (0) | 2022.07.17 |



